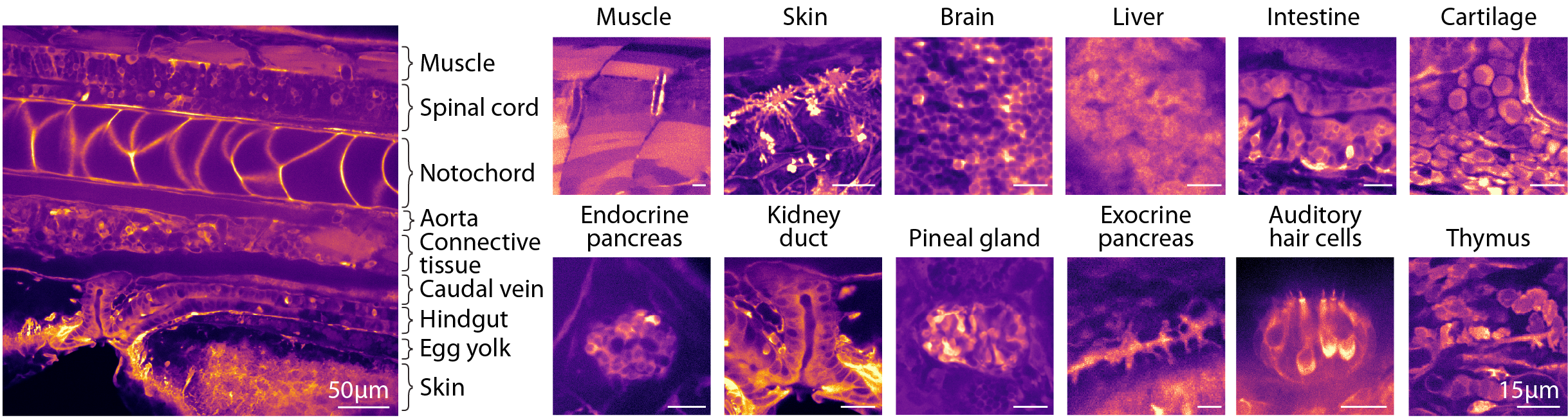
Despite ubiquitous expression in tightly packed cells, we found that tissues could be distinguished by their distinct visual textures arising from variations in cell morphologies, baseline calcium, and sensor expression levels. Together, this enables the recognition of the main organs and bodily tissues, including muscle, liver, kidney, brain, and intestine, as well as more anatomically confined tissues such as the pineal gland and the endocrine pancreas. For more granular identification at the level of cell types, we later introduce Whole-Body Expansion Microscopy.
The identification of all areas was achieved by comparing with established anatomical references, including histological and electron microscopy databases1,2,3,4. A minority, approximately ~10% of other regions, exhibit characteristic texture but remain more difficult to classify.
1. Comparative analysis of fixation and embedding techniques for optimized histological preparation of zebrafish, Comparative Biochemistry and Physiology Part - C: Toxicology and Pharmacology, 2018, Copper, Jean E. and Budgeon, Lynn R. and Foutz, Christina A. and van Rossum, Damian B. and Vanselow, Daniel J. and Hubley, Margaret J. and Clark, Darin P. and Mandrell, David T. and Cheng, Keith C.
2. The zebrafish: atlas of macroscopic and microscopic anatomy, , 2013, Holden, Joseph A and Layfield, Lester L and Matthews, Jennifer L
3. Whole-brain serial-section electron microscopy in larval zebrafish, Nature, 2017, Hildebrand, David Grant Colburn and Cicconet, Marcelo and Torres, Russel Miguel and Choi, Woohyuk and Quan, Tran Minh and Moon, Jungmin and Wetzel, Arthur Willis and {Scott Champion}, Andrew and Graham, Brett Jesse and Randlett, Owen and Plummer, George Scott and Portugues, Ruben and Bianco, Isaac Henry and Saalfeld, Stephan and Baden, Alexander David and Lillaney, Kunal and Burns, Randal and Vogelstein, Joshua Tzvi and Schier, Alexander Franz and Lee, Wei Chung Allen and Jeong, Won Ki and Lichtman, Jeff William and Engert, Florian
4. Spatial and temporal expression of the zebrafish genome by large-scale in situ hybridization screening., Methods Cell Biol, 2004, Thisse, B. and Heyer, V. and Lux, A. and Alunni, V. and Degrave, A. and Seiliez, I. and Kirchner, J. and Parkhill, JP. and Thisse, C.