Registration
We developed a registration algorithm based on iterative optical flow estimation1 that estimates motion patch-wise and combines smoothness regularization, signal-to-noise weighting of gradients, and multi-resolution registration to favor convergence to an optimal tissue alignment for each time point. This achieved high-quality alignment, and most cells were accurately registered throughout the experiments. To evaluate registration accuracy, we employed fiduciary landmarks provided by a sparse transgenic line Tg(phox2bb:eGFP)2, used solely for assessment. Although at times the rapid contractions of gastrointestinal tissues posed challenges, most cells were well-registered for almost all time points. Finally, a registration reliability map is applied to exclude specific subregions at time points with uncertain registration accuracy.
Registration alogrithm
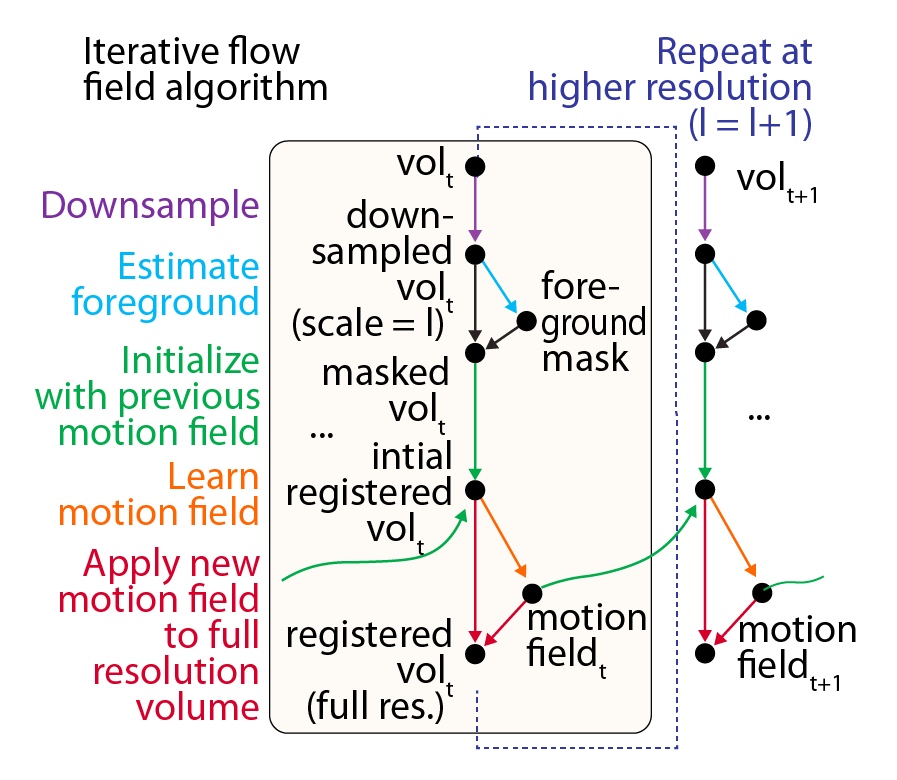
Workflow for WBI data registration utilizing multi-scale iterative estimation of local motion fields, with example results demonstrating temporal alignment of the tissue.
Registration results
Sagittal view of the registration channel derived from Tg(β-actin:mCherry-CAAX) line, imaged with 20× 0.75 NA air objective. Left: data pre-registration; Right: data post-registration.