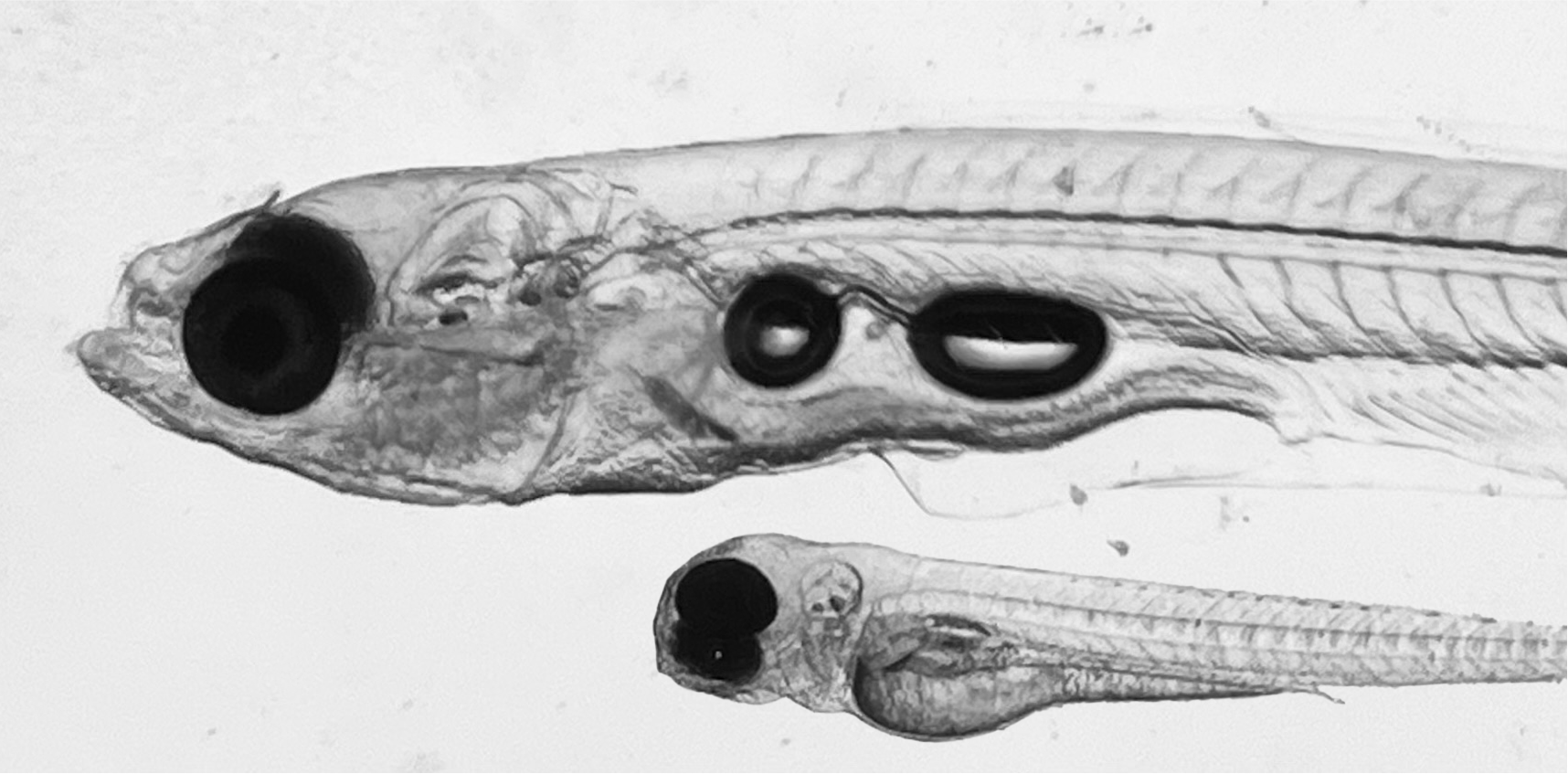
The transparency of young zebrafish and adult danionella species allow for optical access to the inside of these vertebrates.
Brightfield image of a young Danio rerio and a mature Danionella cerebrum.
Transgenic animals are engineered to express fluorescent sensors in all cells.
Fluorescence image of transgenic animal expressing the fluorescent indicator GCaMP7f in all cells of its body.
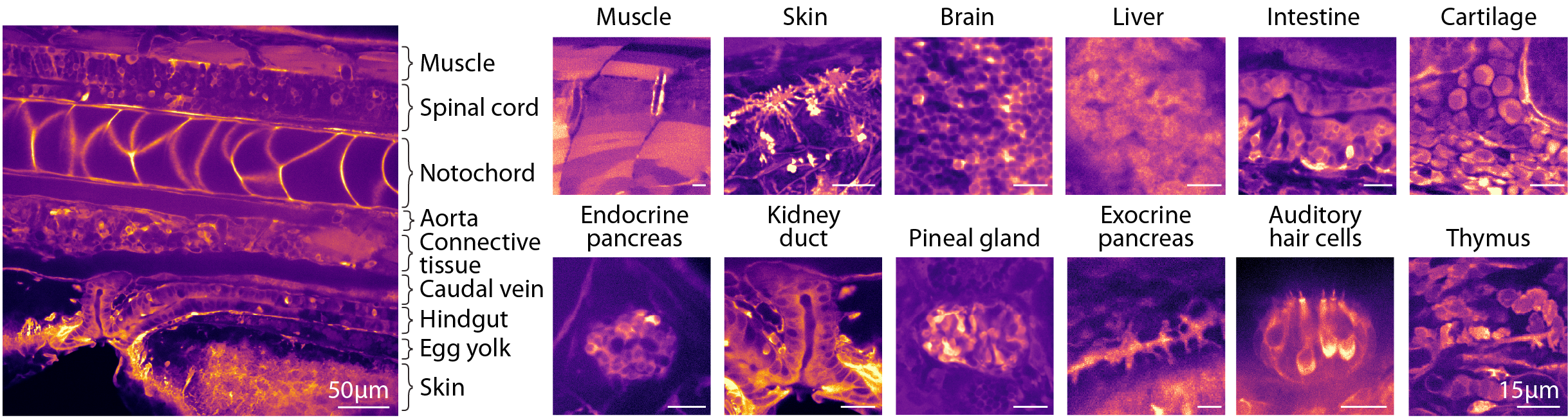
Characteristic visual appearance of organs allows for label-free tissue identification.
Images showing examples of identifiable tissues each displaying distinct visual textures.
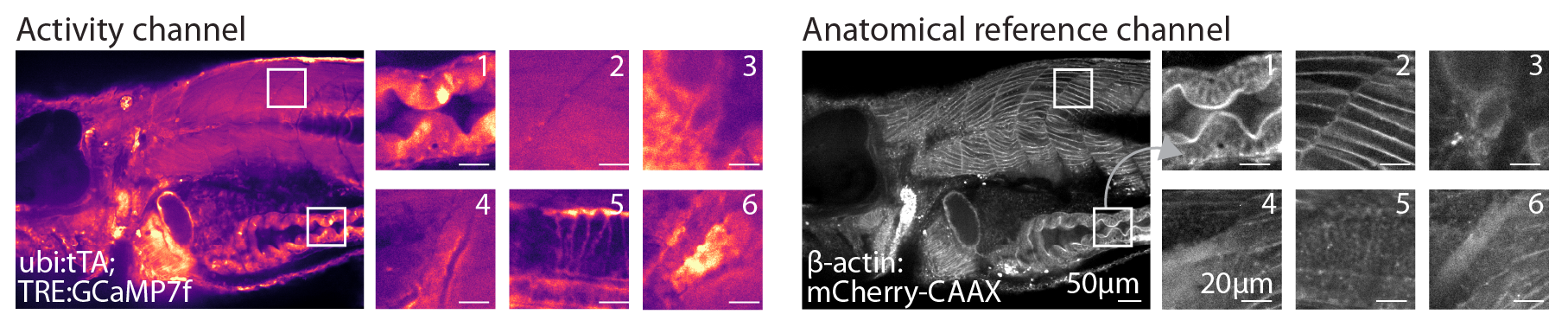
Simultaneously imaging an anatomical reference channel is critical to being able to subsequently reigster WBI data.
Example plane of anatomical reference channel (left) and physiological channel (right). The anatomical channel is based on a transgenic line what expresses a membrane-targeted fluorescent in all cells providing high-frequency texture across the body.
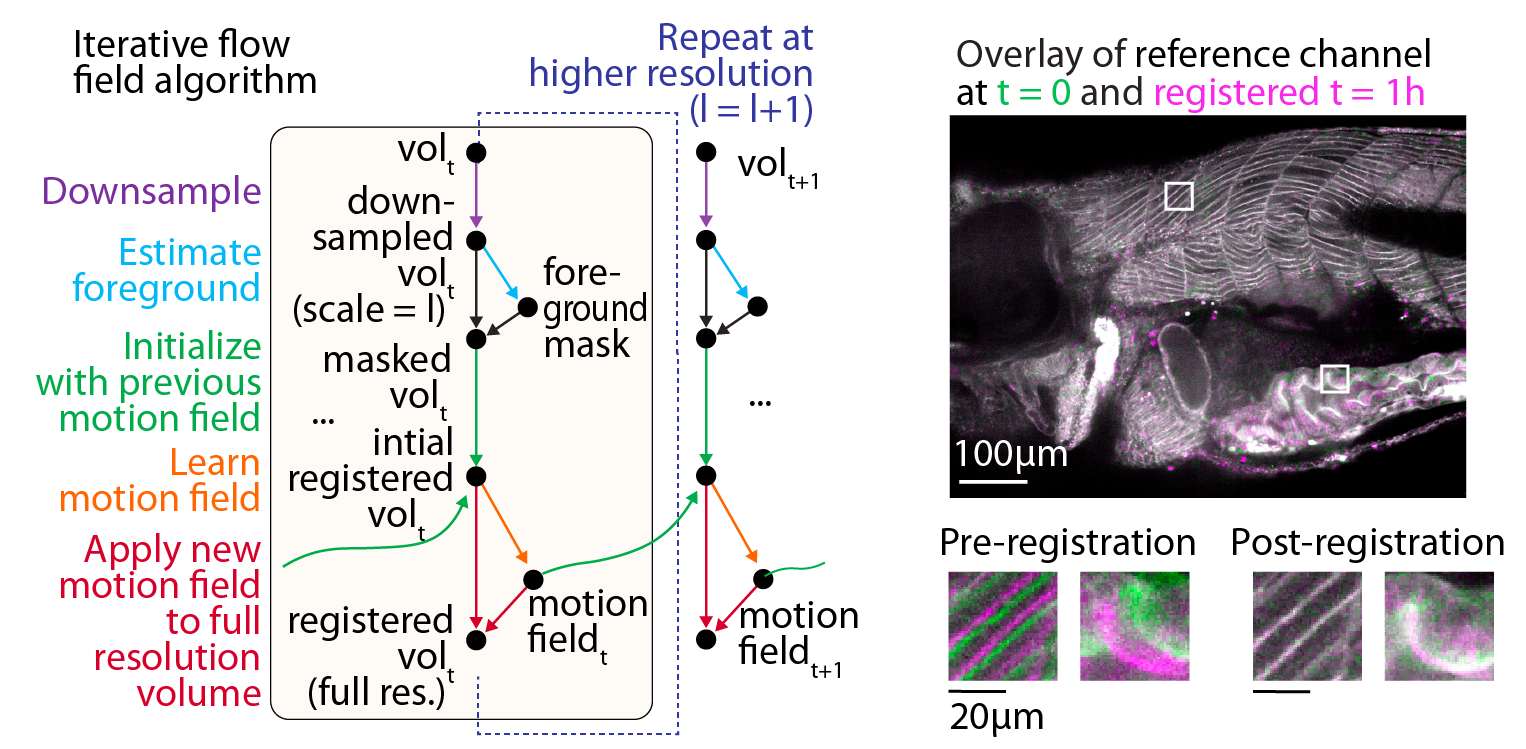
The WBI registration algorithm is based on iterative optical flow estimation and estimates motion patch-wise, combining smoothness regularization, signal-to-noise weighting of gradients, and multi-resolution registration to favor convergence to an optimal tissue alignment for each time point.
Workflow for WBI data registration utilizing multi-scale iterative estimation of local motion fields, with example results demonstrating temporal alignment of the tissue.
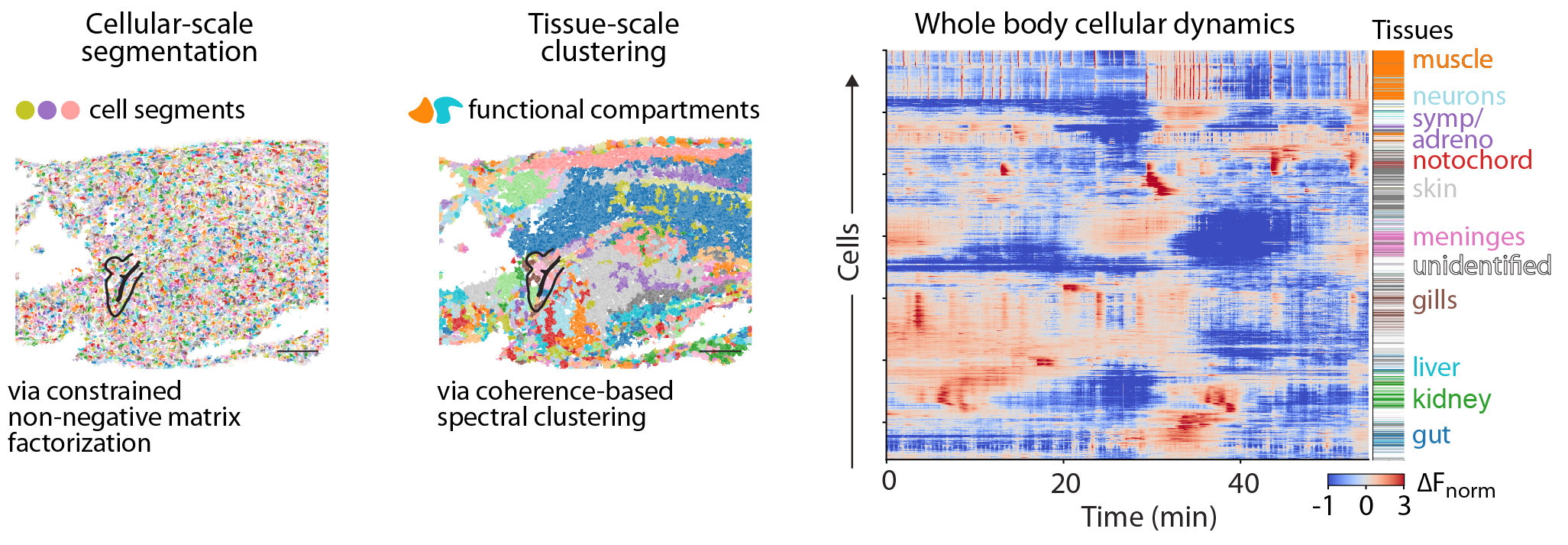
To transform the registered data into cellular-scale activity traces and relate these to organs, tissues, and cell types, we use 1) an algorithm based on constrained non-negative matrix factorization to segment the data into cellular-scale fragments, 2) coherence-based spectral clustering to identify functional tissue compartments.
Example plane showing the weighted spatial footprints of 1) the cell segments derived from a constrained non-negative matrix factorization algorithm, 2) the functional tissue compartments derived from coherence-based spectral clustering.
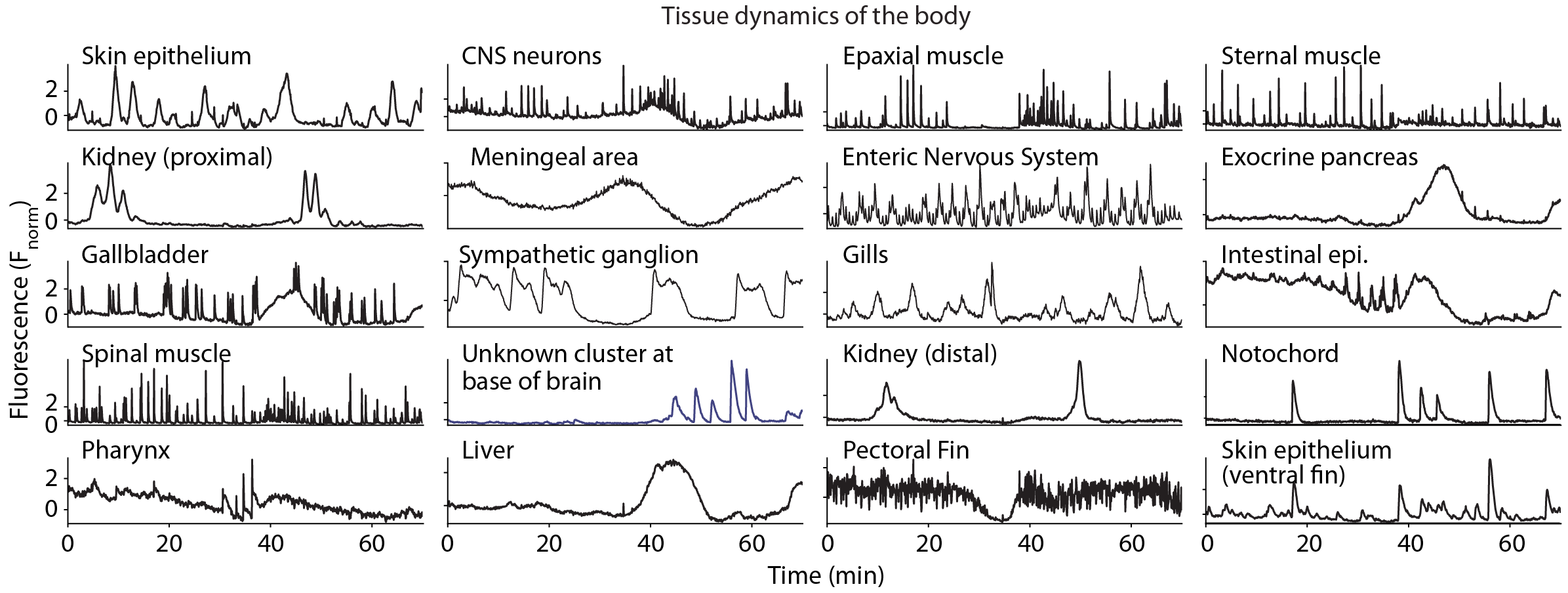
Cells throughout the body exhibit a range of calcium dynamics, showcasing diverse temporal spectra, including oscillatory, pulsatile, and bistable activity. Critically these all fall within the dynamics range of standard fluorescent sensors (e.g., GCaMP7f). Cell types exhibit characteristic calcium dynamics, which can be leveraged to organize the data into interpretable and readily identifiable clusters at the tissue level.
Examples of distinct dynamic profiles observed across functional tissue compartments, including high-frequency dynamics, slower evolving activity, tonic bi-stable dynamics as well as oscillatory activity.
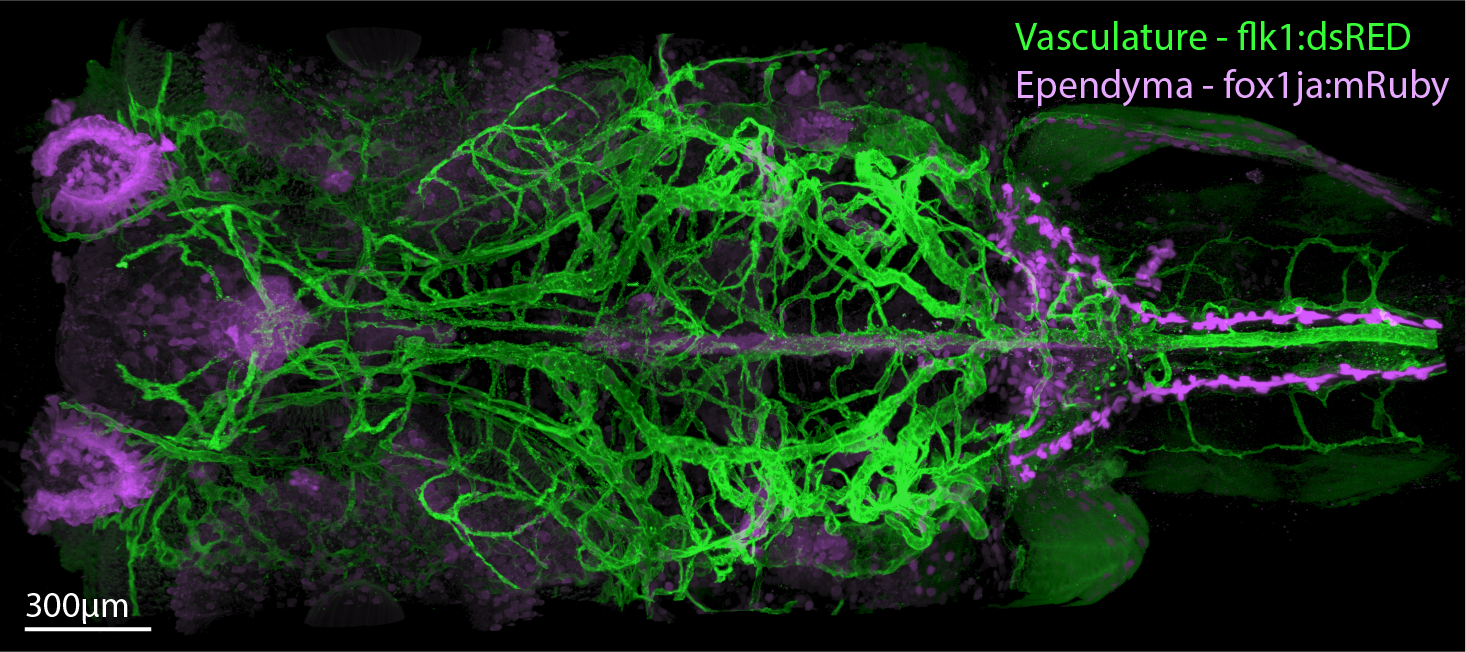
Interpreting and following up WBI observation requires having knowledge of tissue morphology and cellular identity. To acquire this, we developed an advanced enzyme-free, rapid, and robust Whole-Body Expansion-Microscopy (WB-ExM) protocol that enables the acquisition of molecular and cell-type information at subcellular resolution throughout entire larval and juvenile zebrafish and mature Danionella, achieving up to 5× expansion.
Dorsal section of double transgenic sample labeling the ventricular and vascular systems expanded 2x.
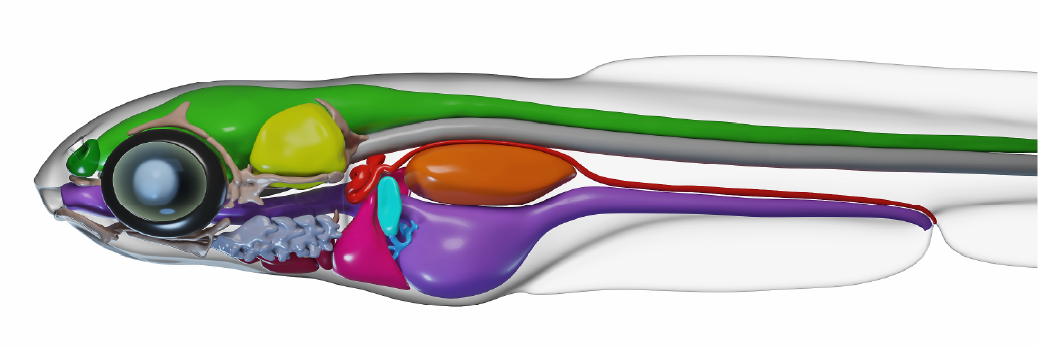
We build a 3D bio-realistic model using WB-ExM to build a reference model with which to cross-reference functional findings.
3D bio-realistic model generated from an ExM-derived meshes and assembled and rendered in Blender
WBI enables drug screens in disease models that allow for tracking the effects at multiple spatial scales from cells to the whole body and at temporal scales from seconds to days.
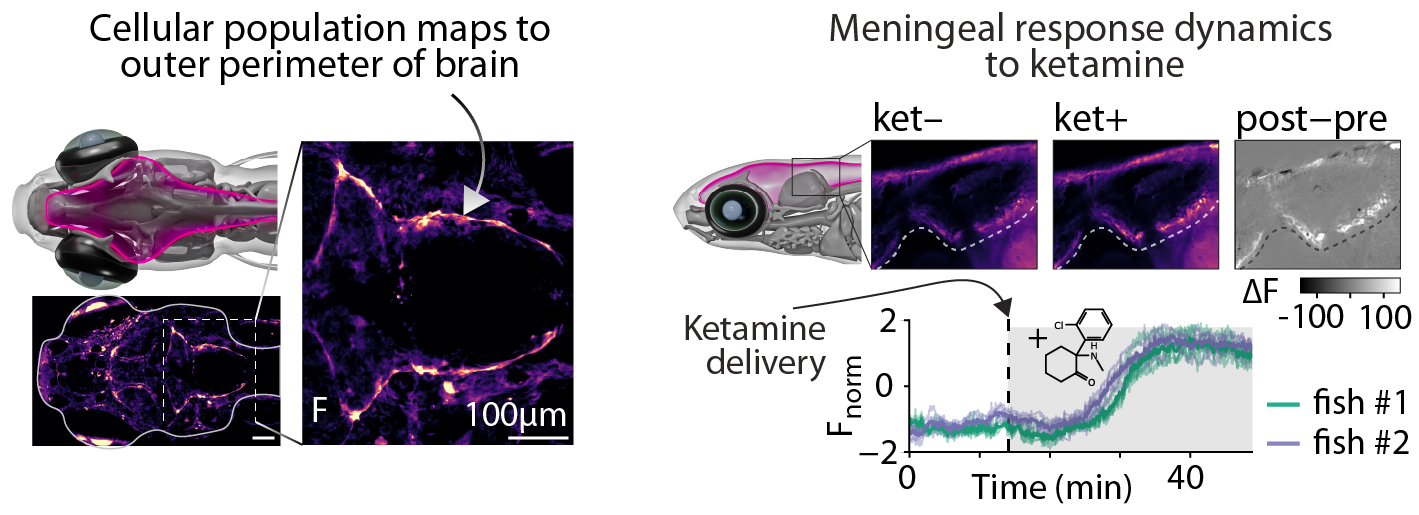
Meningeal response to ketamine exposure. Animals were exposed to ketamine following a 25 min period of baseline imaging. Top) Sagittal section showing pre- and post-exposure to ketamine; Bottom) activity traces from meningeal regions.
WBI enables to screen for stimulus responses, thereby uncovering new cellular properties.
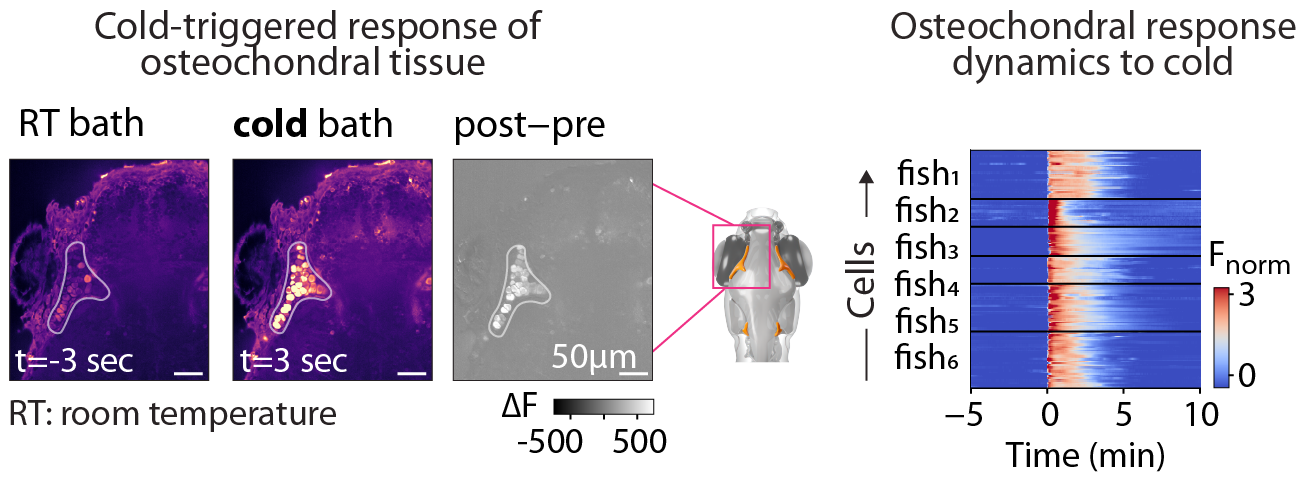
WBI screen for cold-responsive cells. Transverse section of the anterior portion of the cranium shown pre- and post-exposure to cold. Raster of Ca2+ levels in osteochondral cells aligned to cold stimulus onset, showing time-locked responses in 6 animals.